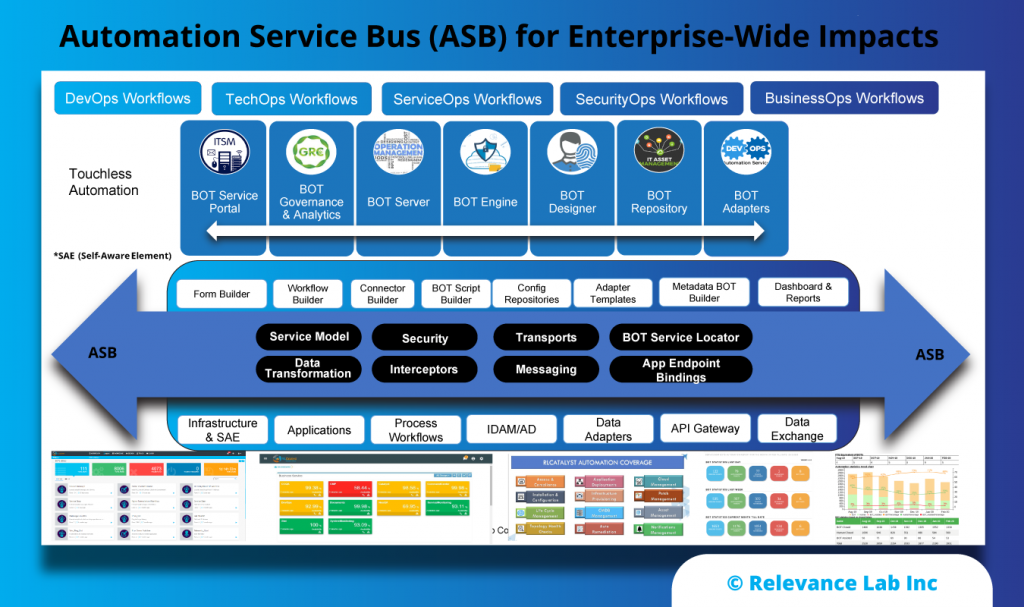
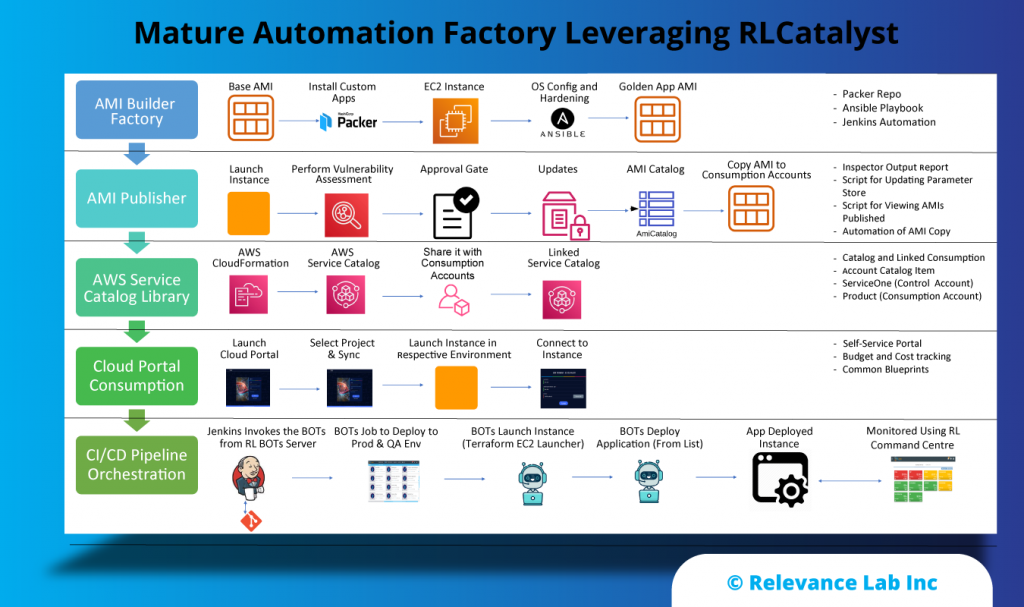
2022 Blog, DevOps Blog, Blog, Featured
Automated deployment of software makes the process faster, easier, repeatable, and more supportable. A variety of technologies are available for deployment, but you need not necessarily choose a complex automation approach to reap the benefits. In this blog, we will cover how Relevance Lab approached using automation for the deployment of their RLCatalyst Research Gateway solution.
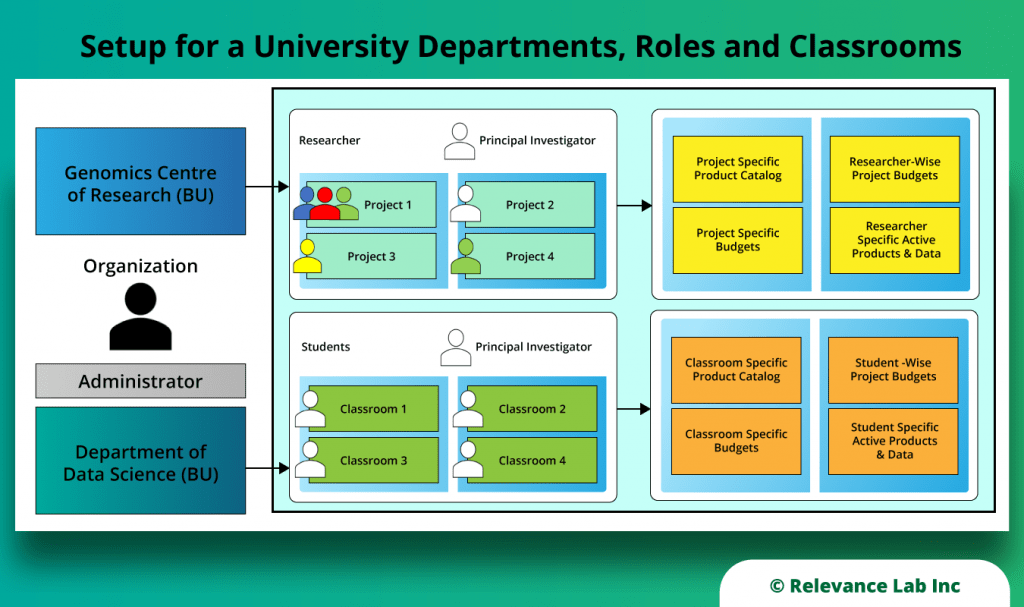
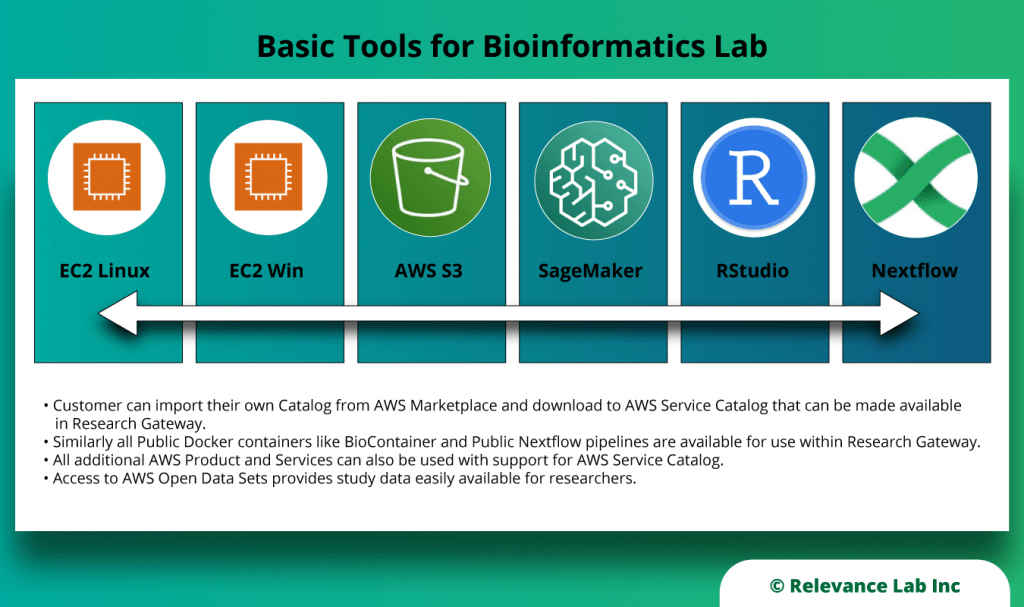
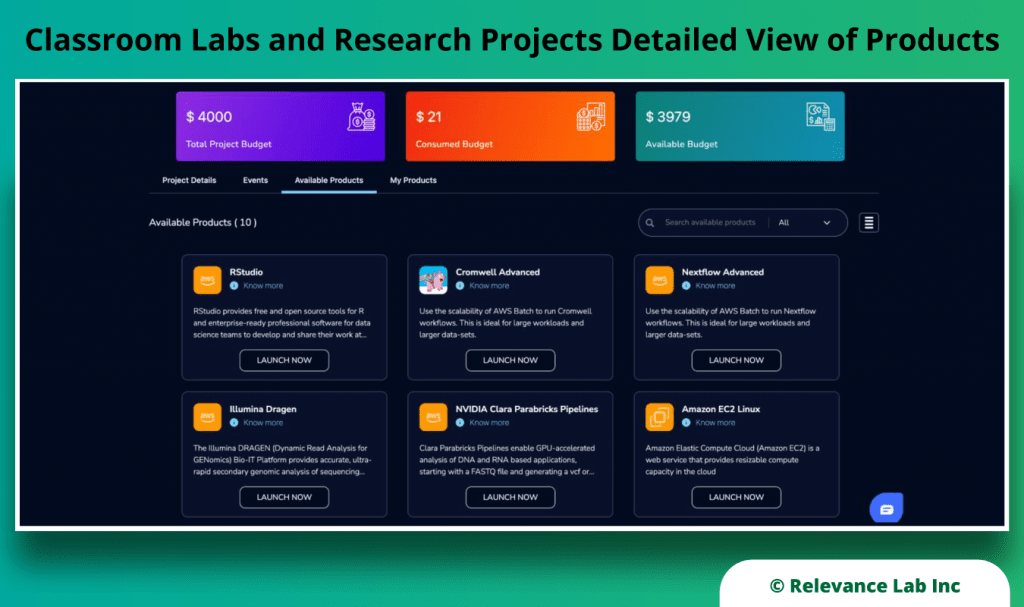
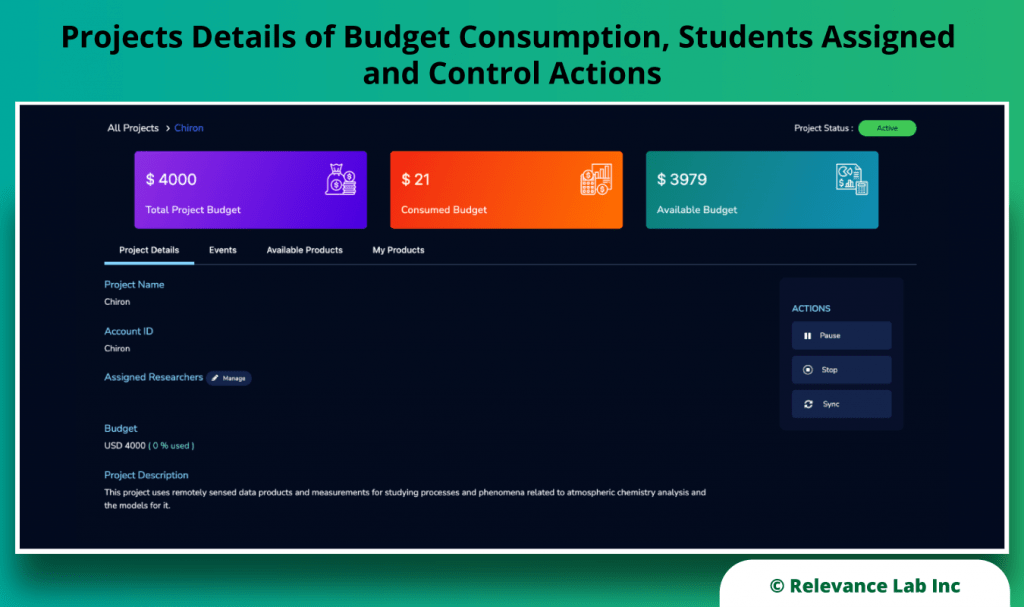
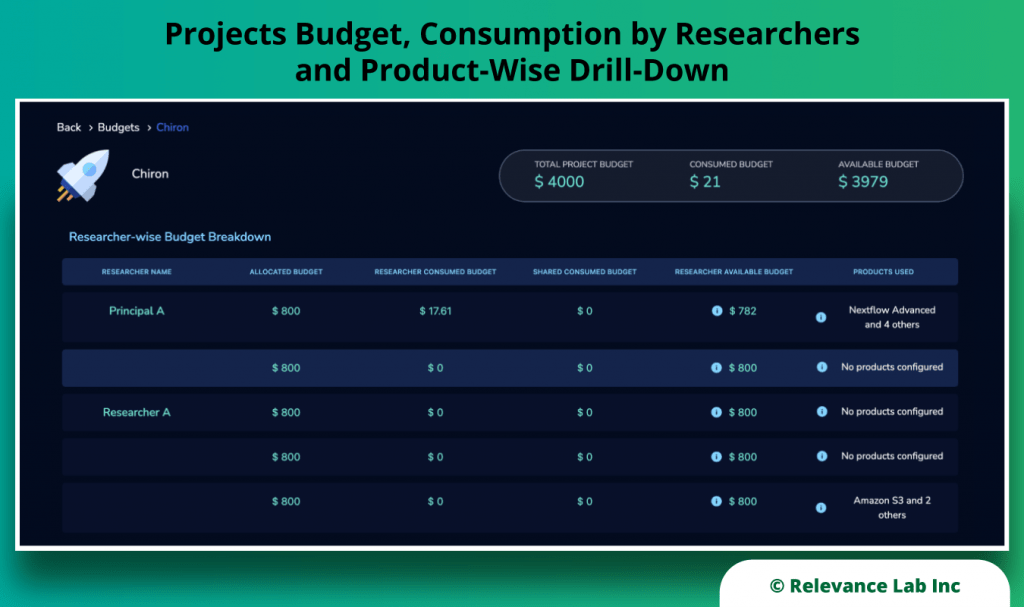
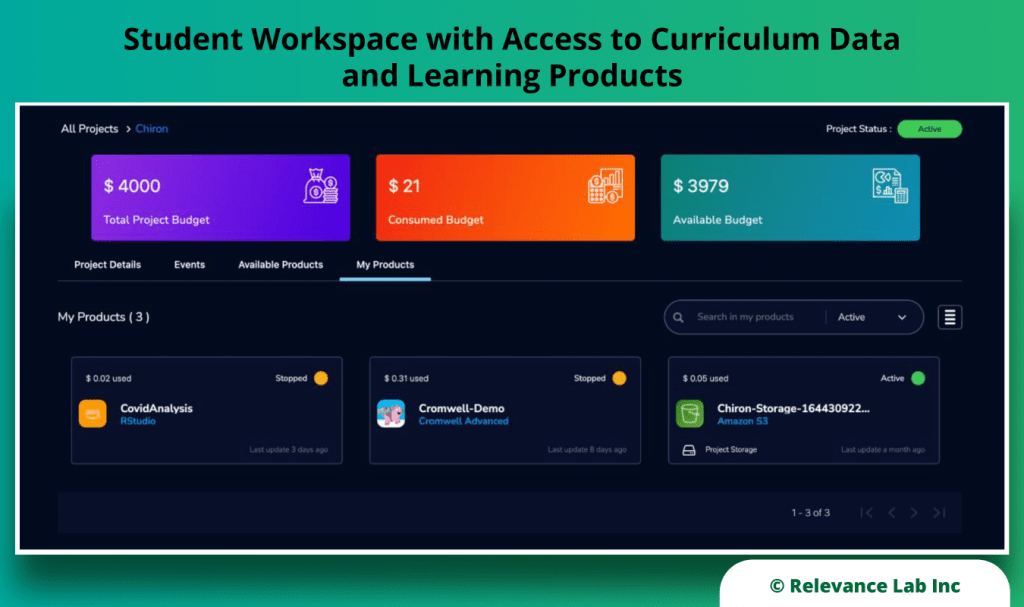
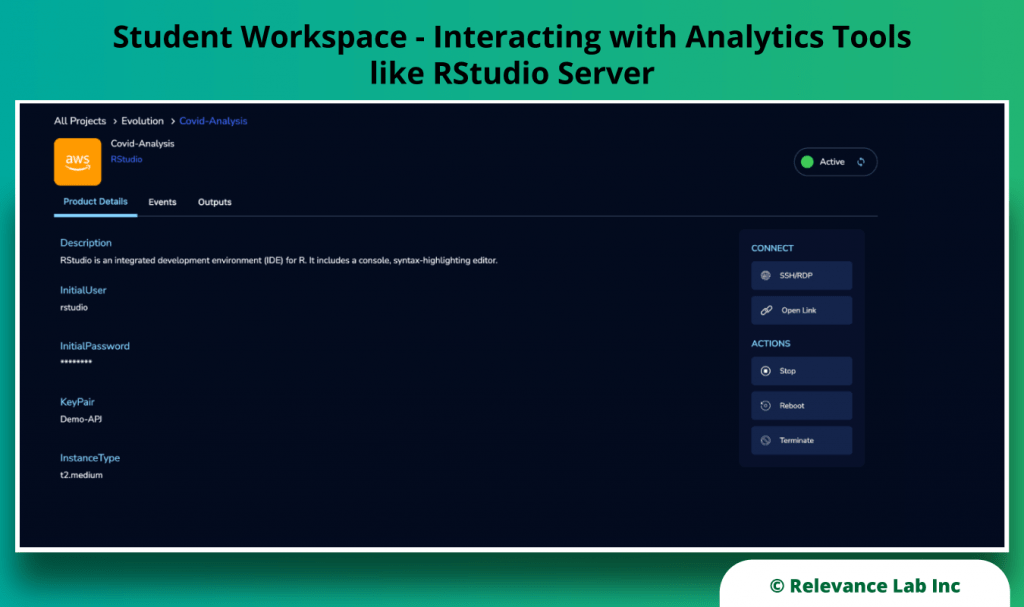
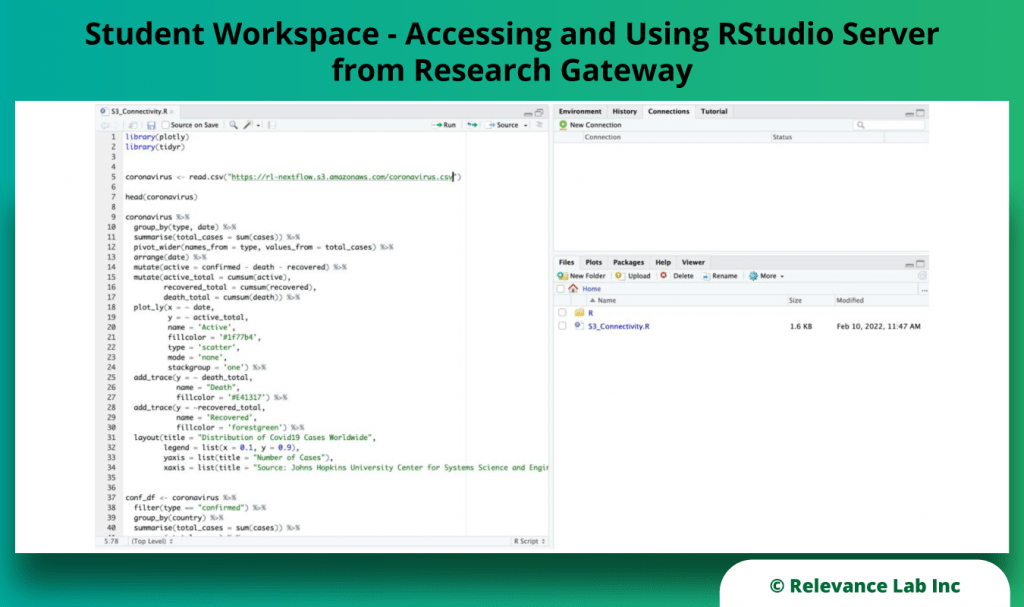
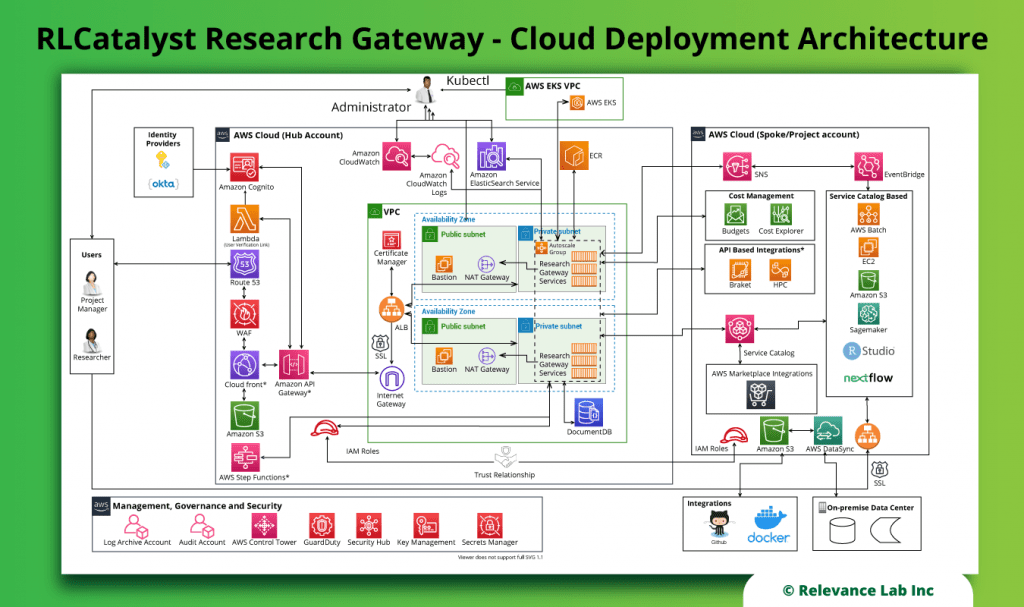
RLCatalyst Research Gateway solution from Relevance Lab provides a next-generation cloud-based platform for collaborative scientific research on AWS with access to research tools, data sets, processing pipelines, and analytics workbenches in a frictionless manner. The solution can be used in the Software as a Service (SaaS) mode, or it can be deployed in customers’ accounts in the enterprise mode. It takes less than 30 minutes to launch a working environment for Principal Investigators and Researchers with security, scalability, and cost governance.
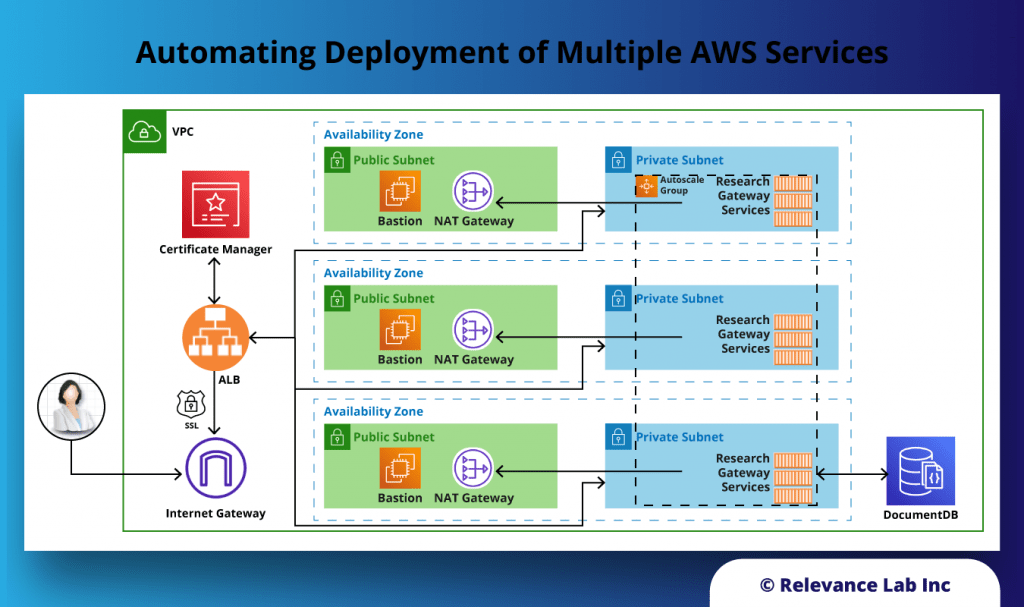
During the deployment of this solution, several AWS resources are created:
- Networking (VPC, Public and private subnets, Internet and NAT Gateways, ALB)
- Security (Security Groups, Cognito Userpool for authentication, Identity and Acess Management (IAM) Roles and Policies)
- Database (AWS DocumentDB cluster)
- EC2 Compute
- EC2 Image Builder pipelines
- S3 Buckets (storage)
- AWS Service Catalog products and portfolios
When such a variety of resources are to be created, there are several benefits of automating the deployment.
- Faster Deployment: It takes an engineer at least a few hours to deploy all the resources manually, assuming everything works to plan. If errors are encountered, it takes longer. With an automated deployment, the process is much quicker, and it can be done in 15-30 minutes.
- Easier: The deployment automation encapsulates and hides a lot of the complexity of the process, and the engineer performing the task does not need to know a lot of the different technologies in depth. Also, since the automation has been hardened over time through repeated testing in the lab, much of the error handling has been codified within the scripts.
- Repeatable: The deployment done via automation always comes out exactly as designed. Unlike manual deployment, where unforced user errors can creep in, the scripts perform each run exactly the same. Also, scripts can be coded to fix broken installs or redeploy solution software.
- Supportable: Automation scripts can have logging, which makes it easy for support personnel to help in case things don’t go as planned.
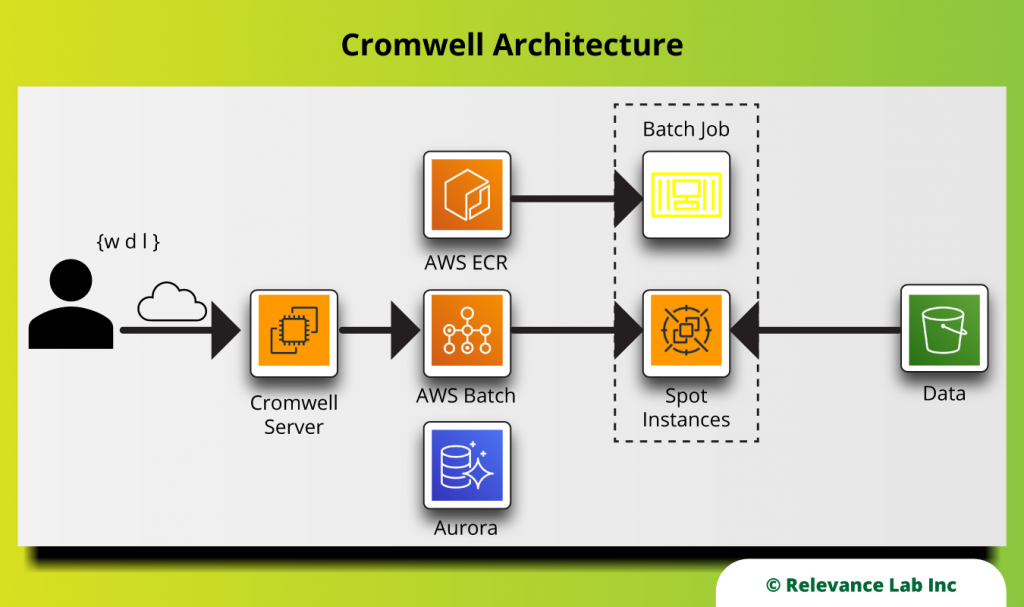
There are many technologies that can help automate the deployment of software. These include tools like Chef and Ansible, language-specific package managers like PyPI or npm, and Infrastructure as Code (IaC) tools like CloudFormation or Terraform. For RLCatalyst Research Gateway, which is built on AWS, we picked CloudFormation Templates (CFT) for our IaC needs in combination with plain old shell scripts. Find our deployment scripts on Github.
- Pre-requisites: We deploy Research Gateway in a standard Virtual Private Cloud (VPC) architecture with both public and private subnets. This VPC can be created using a quickstart available from AWS itself.
- Infrastructure: The infrastructure is created as five different stacks.
- Amazon S3 bucket: This is used to hold all the deployment artifacts like CFT templates.
- AWS Cognito UserPool: This is used for authentication.
- AWS DocumentDB: This is used to store all persistent data required by Research Gateway.
- Amazon EC2 Image Builder: Pipelines are created to rebuild Amazon Machine Image (AMI) for the standard catalog items that are AMI-based. This ensures that the AMIs have the latest patches and security fixes.
- Amazon EC2 (main stack): This hosts the Research Gateway portal.
- Configuration: Some of the instance-specific data is part of the configuration, which is stored in one of the following ways.
- Files: Configuration files are created during the deployment process, using data provided at the time. These files are referred by the solution software to customize its behavior. File-based configurations are easier to access for support personnel and can be easily checked in case the solution software is not behaving as expected.
- Database Entries: A configs collection in the database hosts some of the information. Ideally, all configurations can reside in the database, but because the database is encrypted and has restricted access, we prefer to keep some of the configurations outside the DB.
- AWS Systems Manager (SSM) Parameter Store: Some configurations, especially those related to AMIs, which are resolved by CFTs at run-time, are maintained in the AWS SSM Parameter store.
- Research Gateway Solution Software: Distributed as docker images via AWS Elastic Container Registry (ECR). This allows us to distribute the solution software privately to the customers’ AWS accounts. Our solution software runs as a set of docker services. A variation of the deployment script can also deploy this as services into AWS Elastic Kubernetes Service.
- Load-balancing: The EC2 instances deployed register themselves with Target Groups, and an Application Load Balancer serves the application securely over SSL using certificates hosted in AWS Certificate Manager.
Once the solution software is deployed, and the portal is running and reachable, the first user (an Admin role) is created using a script. Using that Administrator user credentials, the rest of the onboarding process can be completed by the customer from the UI.
Summary
Using the automated deployment process, an instance of the RLCatalyst Research Gateway can be provisioned and configured in less than 30 minutes. This allows customers to start using the solution quickly and derive maximum benefits from their investment with minimum effort.
If you would like to launch your scientific research environment in less than 30 minutes with RLCatalyst Research Gateway or would like to learn more about it, write to us at marketing@relevancelab.com.
References
Architecting a Cloud-based Application with AWS Best Practices
Enabling Frictionless Scientific Research in the Cloud with a 30 Minutes Countdown Now!