2023 Blog, AWS Platform, Blog, Featured, Feature Blog
Relevance Lab (RL) has been an AWS (Amazon Web Services) partner for more than a decade now. While the journey started as a Services Partner it has now extended and matured to a niche technology partner with multiple solutions being offered on AWS Marketplace.
Here is a Quick Snapshot of AWS Capabilities:
- RL is involved in Plan-Build-Run lifecycle of Cloud adoption by enterprises over a multi-year transformation journey.
- The approach to Cloud Adoption is built on some key best practices covering Automation-First Approach, DevOps, Governance360, and Application-Centric Site Reliability Engineering (SRE) focus.
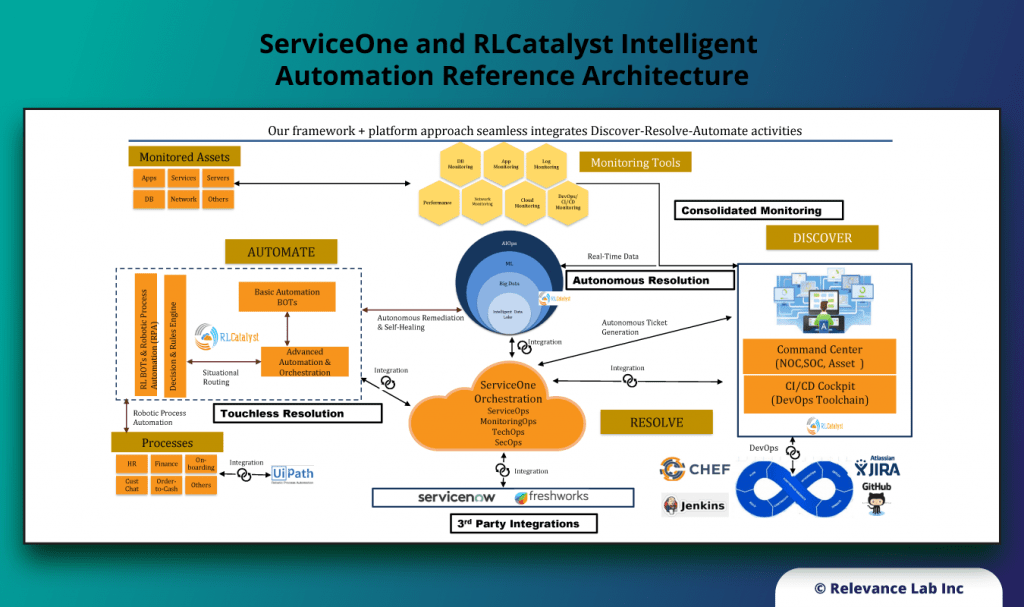
- In Cloud Managed Services we cover all aspects of DevOps, AIOps, SecOps and ServiceDesk Ops leveraging our Automation Platforms – RLCatalyst BOTs, Command Centre, ServiceOne.
- Involved with 50+ Cloud engagements covering large scale (5000+ nodes, 15+ regions, 200+ apps, 5.0+M annual spends) setups and optimization.
- Deep partnership with AWS and ServiceNow to bring end-to-end Governance360 covering Asset Management, CMDB, Vulnerability & Patch Management, SIEM/SOAR, Cost/Security/Compliance Dashboards.
- Products created and deployed on AWS for Self-Service Cloud Portals and Purpose-built cloud solutions covering HPC (High Performance Computing), Containers, Service Catalog, Cost & Budget tracking, and Scientific Research workflows.
- Our work and resources cover Cloud Infrastructure, Cloud Apps, Cloud Data and Cloud Service Delivery with 800+ cloud trained resources, 450+ Cloud specialists and 100+ certifications.
- RL is global number one preferred partner for AWS as an ISV provider for Scientific Research Computing building solutions using AWS Open-Source solutions like Service Workbench.
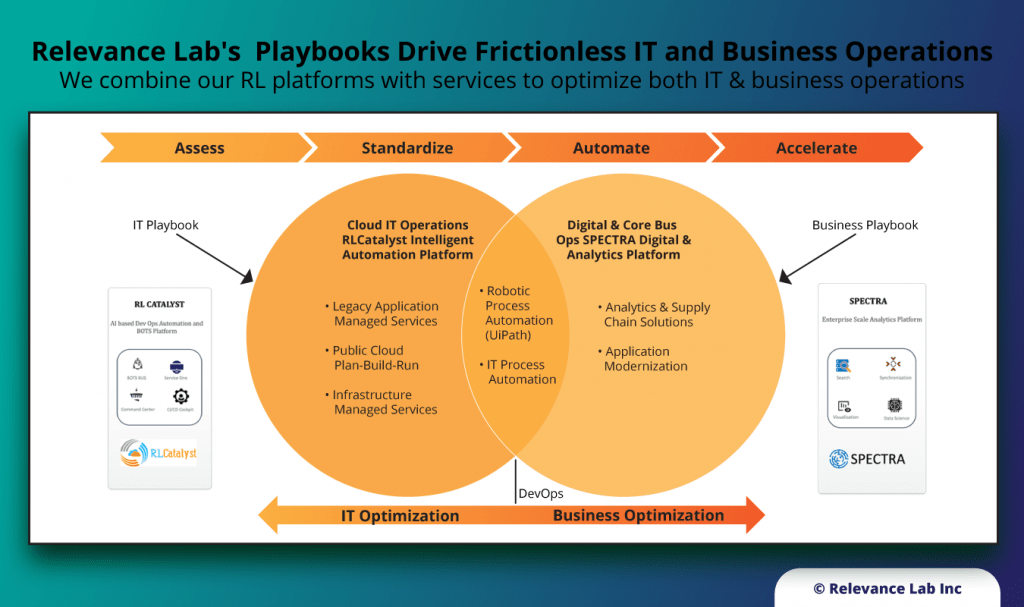
Our unique positioning of Products + Services helps create platform-based offerings delivered as playbooks for digital transformation.
Our key focus areas in Cloud Offerings are the following:
- Cloud Management & Governance
- Full Lifecycle Automation and Self-Service Portals
- Containers, Microservices, Well Architected Frameworks and Kubernetes
- AIOps and Site Reliability Engineering
What Makes Us Different?
- Automation-First approach across “Plan, Build & Run” Lifecycle helps customers use “Cloud the Right Way” focused on best practices like “Infrastructure as a Code” and “Compliance as a Code.”
- RLCatalyst Products offer Enterprise Cloud Orchestration and Governance with a pre-built library of quick-starts, BOTs, Self-Service Cloud Portals, and Open-source solutions.
- AWS + ServiceNow unique specialization leveraged to provide Intelligent Cloud Operations & managed services.
- ServiceOne AIOps Platform covering workload migration, security, governance, CMDB, ITSM and DevOps.
- Frictionless Digital Application modernization and Cloud Product Engineering services for native cloud architecture and competencies.
- Open-Source Co-Development with AWS for Scientific Research Solutions (Higher Ed and Healthcare).
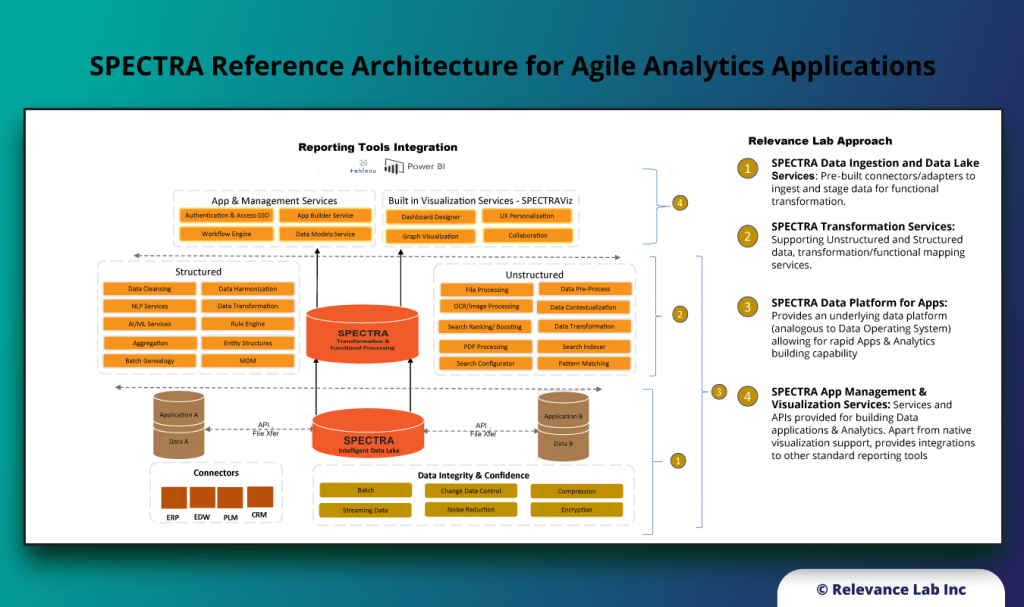
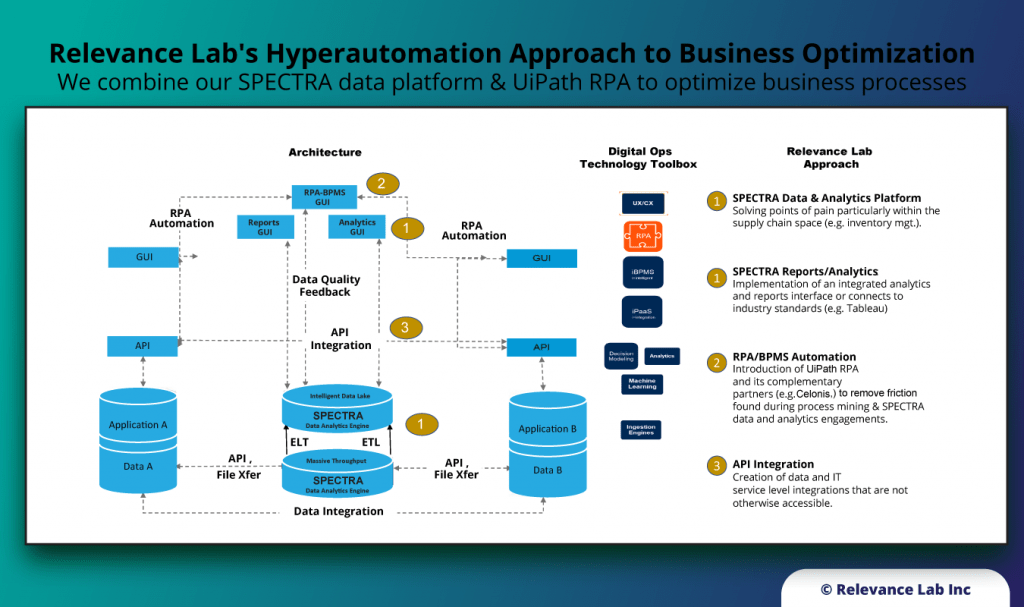
- Agile Analytics with our Spectra Data platform that helps building Enterprise Data Lakes and Supply Chain analytics by with multiple ERP systems connectors.
Our Solutions Sweet Spot
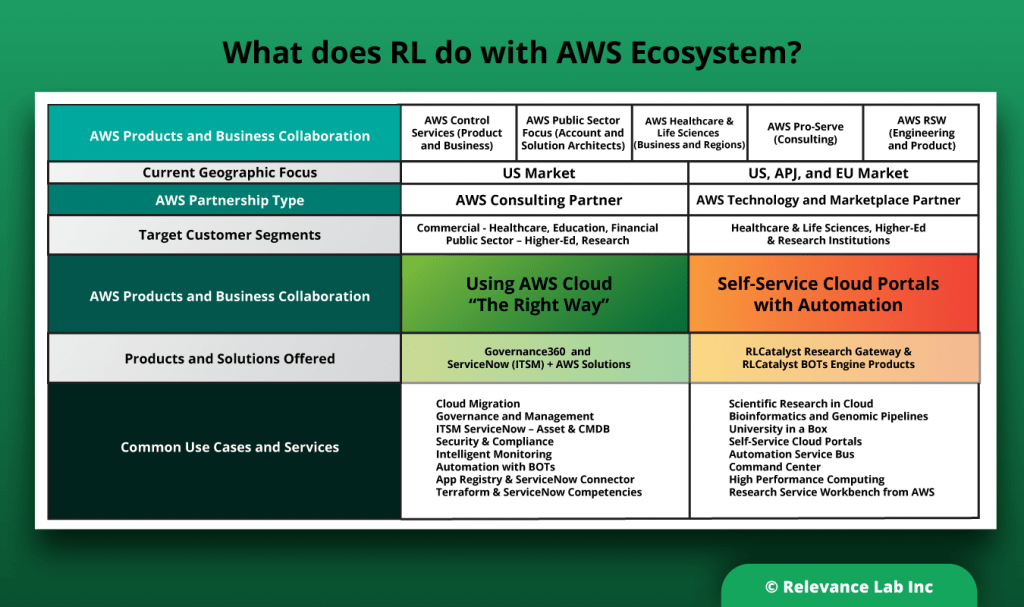
Governance360
Built on AWS Control Services a prescriptive and automated maturity model for proper workload migration, governance, security, monitoring and Service Management.
RLCatalyst BOTS Automation Engine and ServiceOne
Product covering end-to-end automation with a library of 100+ pre-built BOTs. Intelligent user and workspaces onboarding and offboarding.
Research Gateway – Self Service Cloud Portals
Self-Service Cloud Portal for Scientific Research in Cloud with HPC, Genomic Pipelines, covering EC2, SageMaker, S3 etc.
ServiceNow AppInsights built on AWS AppRegistry
Dynamic Applications CMDB leveraging AWS and ServiceNow with focus on Application Centric costs, health, and risks.
DevOps Driven Engineering and Cloud Product Development
DevOps-driven CI/CD, Infra Automation and Proactive Monitoring. AWS Well-architected. Cloud App Modernization, APM, API Gateways, Cloud Integration with Enterprise Systems. AWS Digital Customer Experience competencies
SPECTRA Data Platform for Cloud Data Lakes
Enterprise Data Lake with large data movement from on-prem to Cloud systems and ERP integration adapters for Supply Chain Analytics.
AWS Product Focus Areas
Control Tower, Security Hub, Service Catalog, HPC, Quantum Computing, Data Lake, ITSM Connectors, Well-Architected, SaaS (Software as a Service) Factory, Service Workbench, CloudEndure, AppStream 2.0, QuickStart for HIPPA, Bioinformatics
Focus on Software, Databases, Workloads
Open-source and App development stacks, Java, Python, MS .Net, Cloudera, Databricks, MongoDB, RedShift, Hadoop, Snowflake, Magento, WordPress, Moodle, RStudio, Nextflow
Key Verticals Solutions
- Technology companies (ISVs & startups)
- Media/Publishing/Higher Education/ Research
- Pharma/Healthcare/Life Sciences
- Financial and Insurance
The following are some Customer Solutions highlights:
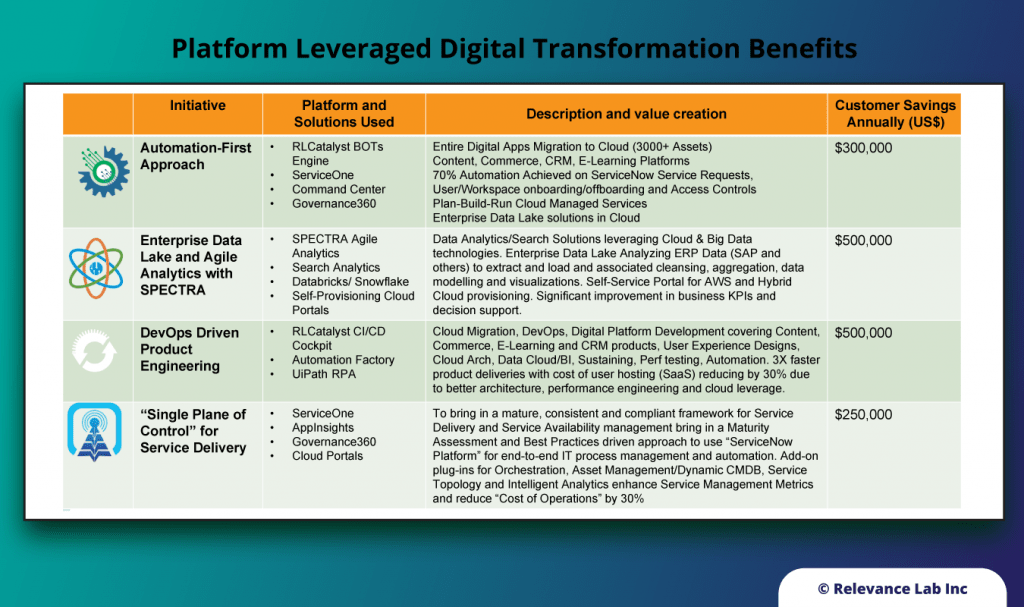
| Digital Publishing & Learning Specialist | Cloud Migration, DevOps, Digital Platform Development covering Content, Commerce, E-Learning and CRM products, User Experience Designs, Cloud Arch, Data Cloud/BI, Sustaining, Perf testing, Automation |
| Global Pharma & Health Sciences Leader | Data Analytics/Search Solutions leveraging Cloud & Big Data technologies. Enterprise Data Lake Analyzing ERP Data (SAP and others) to extract and load and associated cleansing, aggregation, data modelling and visualizations. Self Service Portal for AWS and Hybrid Cloud provisioning |
| Large Financial & Asset Mgmt. Firm | Drive Cloud Adoption, App Modernization and DevOps models as part of IT Transformation journey leveraging their Cloud, Automation and Data Platforms. |
| Specialist Automation ISV | Global partnership working across joint long-term engagements with multiple enterprise customers covering Infrastructure Automation, Application Deployment Automation, Compliance-as-a-Code and Hybrid Cloud Automation. |
Summary
Relevance Lab has close collaboration and partnership with AWS for both products and competencies. We have been part of successful digital transformation with 50+ customers leveraging AWS across Infrastructure, Applications, Data Lakes, and Service Delivery Automation. We enable AWS Cloud adoption “The Right Way” with our comprehensive expertise and pre-built solutions better, faster, and cheaper.
Learn more about our cloud products, services, and solutions, feel free to contact us at marketing@relevancelab.com.
References
Get Dynamic Insights into Your Cloud with an Application-Centric View
Automation of User Onboarding and Offboarding Workflows